5 Read Mapping Genomics Tutorial 2020 2 0 Documentati Vrogue Co

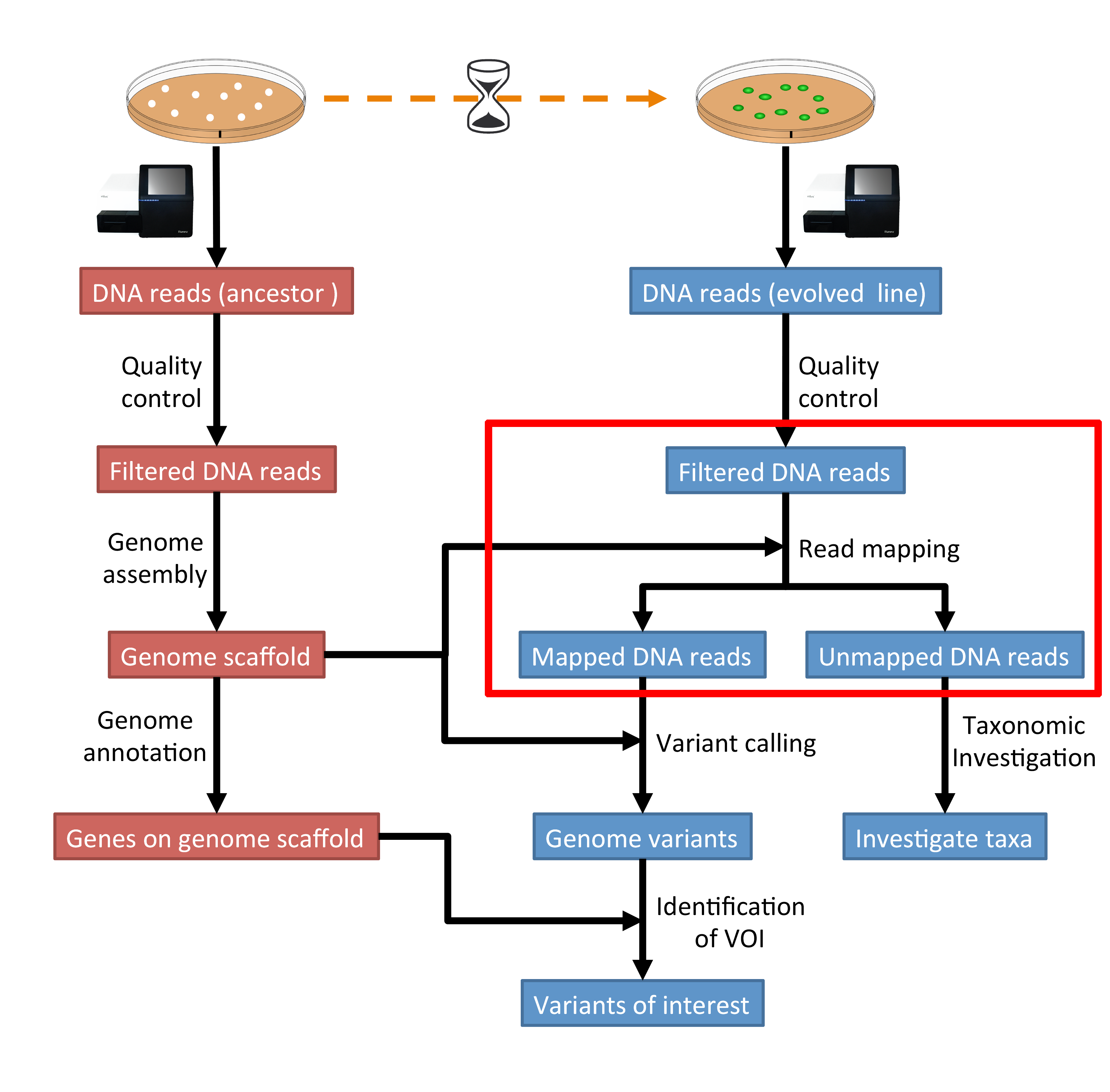
5 Read Mapping Genomics Tutorial 2020 2 0 Documentati Vrogue Co 5. read mapping ¶. 5.1. preface ¶. in this section we will use our skill on the command line interface to map our reads from the evolved line to our ancestral reference genome. note. you will encounter some to do sections at times. write the solutions and answers into a text file. 5.2. This is an introductory tutorial for learning computational genomics mostly on the linux command line. you will learn how to analyse next generation sequencing (ngs) data. the data you will be using is real research data. the final aim is to identify genome variations in evolved lines of bacteria that can explain the observed biological phenotypes.
Sequencing Mapping Vrogue Co In this quality control section we will use our skill on the command line interface to deal with the task of investigating the quality and cleaning sequencing data [kirchner2014]. note. you will encounter some to do sections at times. write the solutions and answers into a text file. 3.2. Genomics tutorial latest 1. introduction; 2. tool installation read mapping: mapping index (bwa) [ dropbox] free document hosting provided by. Figure 4: individual assembly followed by de replication vs co assembly. source: drep documentation. co assembly is more commonly used than individual assembly and then de replication after binning. but in this tutorial, to show all steps, we will run an individual assembly. 4 aug: .ped and .map lines starting with '#' are now consistently treated as comments, following hint #2 in the original ped documentation. we have updated the respective plink 1.9 and 2.0 "file formats" entries, and apologize for overlooking this earlier. 11 dec: ld window cm should work properly now.

Comments are closed.