Single Nucleotide Polymorphism Snp

A Single Nucleotide Polymorphism Snp Download Scientific Diagram In genetics and bioinformatics, a single nucleotide polymorphism (snp snɪp ; plural snps snɪps ) is a germline substitution of a single nucleotide at a specific position in the genome. Snps are genetic variations that occur in a single dna nucleotide. they can act as markers for disease genes or affect gene function. learn more about snps and how they are studied in genomic research.
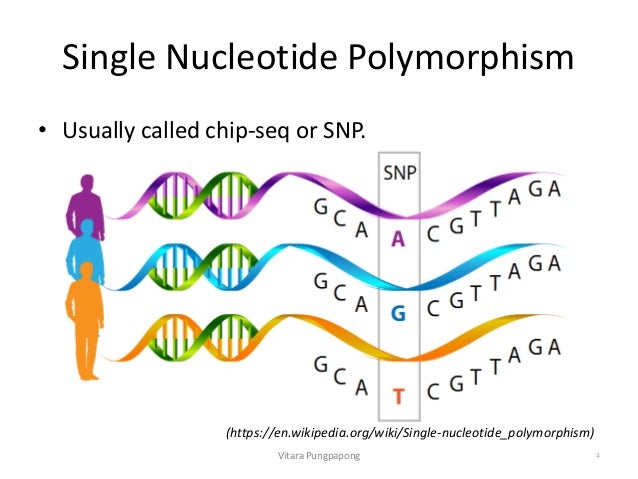
Single Nucleotide Polymorphism Analysis Snps Single nucleotide polymorphism (snp), variation in a genetic sequence that affects only one of the basic building blocks— adenine (a), guanine (g), thymine (t), or cytosine (c)—in a segment of a dna molecule and that occurs in more than 1 percent of a population. A single nucleotide polymorphism (abbreviated snp, pronounced snip) is a genomic variant at a single base position in the dna. scientists study if and how snps in a genome influence health, disease, drug response and other traits. narration. 00:00. … single nucleotide polymorphism, or snp. A single nucleotide polymorphism, or snp, is a single base pair difference in the dna sequence of individual members of a species; not necessarily a pathological mutation, but commonly. The most common variations are single nucleotide polymorphisms (snps), which occur approximately once every 100 to 300 bases. because snps are expected to facilitate large scale association genetics studies, there has recently been great interest in snp discovery and detection.

Single Nucleotide Polymorphism Snp Stock Image C044 6124 A single nucleotide polymorphism, or snp, is a single base pair difference in the dna sequence of individual members of a species; not necessarily a pathological mutation, but commonly. The most common variations are single nucleotide polymorphisms (snps), which occur approximately once every 100 to 300 bases. because snps are expected to facilitate large scale association genetics studies, there has recently been great interest in snp discovery and detection. Single nucleotide polymorphisms (snps), pronounced as “snips,” is the common type of variation found in dna between genes (genetics home reference). each snp differs by a single dna block represented as nucleotide. for example, a snp may be replaced by adenine (a) in place of guanine (g) in a stretch of dna. Single nucleotide polymorphism (snp) refers to specific sites in the human genome where variations can occur, with the possibility of having two or more different nucleotides at a particular position on a chromosome. these variations are common and contribute to human genetic diversity.

Comments are closed.